LMU Newsroom
What is going on at LMU? Everything at a glance in the LMU Newsroom — news, events, interviews, backgrounds, stories.

Team SpiCy on the lookout for oxygen for space missions
Humans can adapt to all kinds of conditions. But how do they get oxygen for space travel? A team of students is working to find a solution.
Read more
AI tool recognizes serious ocular disease in horses
Researchers at the LMU Equine Clinic have developed a deep learning tool that is capable of reliably diagnosing moon blindness in horses based on photos.
Read more
Siegfried in slang
A series of lectures explores how films about the Nibelungen saga have changed over time. We talked to German studies expert Christoph Petersen about an old story and new perspectives.
Read moreINSIGHTS. Magazine

"Really?" - the new issue of INSIGHTS
"Echt jetzt" - the new issue of EINSICHTEN The new edition of the research magazine EINSICHTEN is out, with the main topic: "Really? Is the boundary between natural and artificial increasingly fading?" Click here for the highlights.
Read more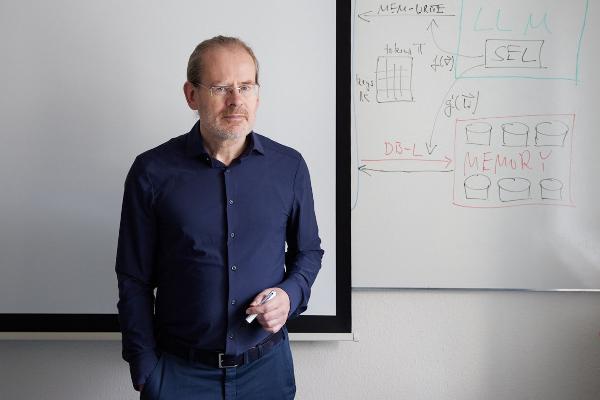
Polyglot machines
How artificial intelligence learns the rich variety of human languages: Hinrich Schütze, computational linguist at LMU, researches multilingual software that can do small languages. From the research magazine EINSICHTEN
Read more
From the steppe to the city
A social transformation is underway in Mongolia, as many nomadic pastoralists move to urban areas. Geographer Lukas Lehnert investigates what this means for the environment and ecosystems. From the research magazine EINSICHTEN
Read more